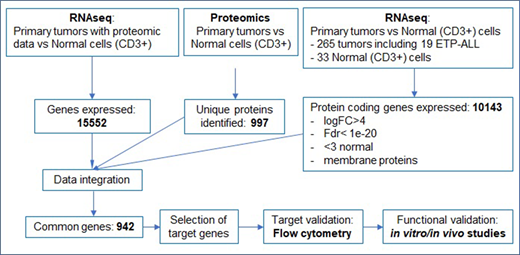
Pediatric T-cell acute lymphoblastic leukemia (T-ALL) is a high-risk disease due to treatment related complications and poor prognosis of patients with relapsed disease. Immunotherapy with monoclonal antibodies (MAbs) and/or chimeric antigen receptor (CAR) T-cells for T-ALL is limited by identification of tumor specific target antigens. Differential expression is necessary to prevent on-target/off-tumor toxicities and fratricide of activated T-cells. Targeting multiple antigens can bypass immune escape and result in improved T-cell effector function, since antigen density correlates with T-cell activation. Here we designed a pipeline (Figure 1) to identify unique surface antigens expressed in T-ALL using proteomic and transcriptomic analyses followed by flow cytometry validation, and functional studies with CAR T cells targeting the identified antigens.
We generated an Illumina total stranded RNAseq library from healthy donor myeloid and lymphoid cells of bone marrow, peripheral blood and cord blood (N= 116). We compared data to 265 St. Jude pediatric T-ALL samples and against 53 normal tissue expression data from the GTEx (Genotype-Tissue Expression) project. To analyze the T-cell surface proteome, we isolated plasma membrane fractions from 11 samples including healthy T-cells and T-ALL cell lines using a differential centrifugation-based method. The purity of the plasma membrane fraction was confirmed by western blot. Na+/K+ ATPase and GAPDH were used as controls for the plasma membrane and cytosolic fractions respectively. Following plasma membrane enrichment, the membrane proteins were applied for proteomic analysis using an advanced TMT-L/LC-MS/MS pipeline, and the acquired proteomic data were further processed via the JUMP software suite. 997 unique proteins were quantified from the membrane fractions. Integrated analysis the transcriptomic and proteomic datasets showed significant correlation and yielded a list of candidate genes, which were validated by flow cytometry on a panel of T-ALL cell lines (CCRF, RPMI8402, and MOLT3) and resting and activated T-cells from healthy donors. We identified GRP78 as one of the differentially expressed cell surface antigens and further confirmed its expression on additional T-ALL cell lines (KE37, PF382, PEER, CEMC7) and 3 PDX samples. Finally, we generated GRP78-CAR T cells and demonstrate that GRP78-CAR T cells recognize and kill GRP78+ T-ALL cells and have potent antitumor activity in xenograft and PDX models.
We have established an unbiased pipeline to identify differentially expressed antigens on the cell surface of T-ALL blasts and created a healthy tissue RNAseq library. The results from our analyses are encouraging and interrogation of our pipeline has yielded differentially expressed immunotherapy targets for the treatment of relapsed refractory T-ALL. Our results highlight the importance of integrated surface proteomics and transcriptomics analysis.
Figure 1: Outline of strategy for target selection:
Hebbar:St. Jude: Patents & Royalties. Epperly:St. Jude: Patents & Royalties. Gottschalk:Inmatics and Tidal: Membership on an entity's Board of Directors or advisory committees; TESSA Therapeutics: Other: research collaboration; Patents and patent applications in the fields of T-cell & Gene therapy for cancer: Patents & Royalties; Merck and ViraCyte: Consultancy. Mullighan:AbbVie, Inc.: Research Funding; Illumina: Consultancy, Honoraria, Speakers Bureau; Pfizer: Honoraria, Research Funding, Speakers Bureau. Velasquez:Rally! Foundation: Membership on an entity's Board of Directors or advisory committees; St. Jude: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal